Documentation
Getting Started
GRNsight is a web application for visualizing models of small- to medium-scale gene regulatory networks (GRNs). GRNsight was created to work with the input and output spreadsheets used by GRNmap, a MATLAB program that estimates the parameters and performs forward simulations of a differential equations model of a GRN. However, GRNsight can be used to visualize any medium-scale GRN that is specified by the appropriate spreadsheet format. GRNsight is best-suited for visualizing networks of fewer than 35 genes (nodes) and 70 regulatory relationships (edges), but can display networks of up to 75 nodes and 150 edges. Although originally designed for gene regulatory networks, we believe that GRNsight has general applicability for displaying any small, unweighted or weighted network with directed edges for systems biology or other application domains. The following sections describe how to use GRNsight.
GRNsight has been tested with and confirmed to be working in Chrome version 105.0.5195.127 or higher and Firefox version 105.0 or higher on Windows 7 and Mac OS X operating systems. It may not work with other browsers, versions, or operating systems.
-
There are three ways to load a network into GRNsight:
- You can load a network from one of our available Demo files, as explained in Section 1a.
- You can create and load a GRNsight Compatible Input Workbook, as explained in Section 1b.
- You can create and load a Network from genes stored in our Network Database, as detailed in Section 1c.
If you do not have your own input spreadsheet, you can load and view demonstration gene regulatory networks.
- Selecting the menu item Demo > Demo #1: Unweighted GRN (15 genes, 28 edges, Dahlquist Lab unpublished data) or Demo #3: Unweighted GRN (21 genes, 31 edges) will load a network graph with black edges and regular (pointy) arrowheads. Selecting the menu item Demo > Demo #2: Weighted GRN (15 genes, 28 edges, Dahlquist Lab unpublished data) or Demo #4: Weighted GRN (21 genes, 31 edges, Schade et al. 2004 data) will load a network graph with colored edges of varying thicknesses. You can access demo selection menu in the Network side-bar menu item and the Network menu item. See Section 2 for details on how GRNsight displays the graph.
- If you have a Microsoft Excel Workbook file (*.xlsx) that was used as an input or generated as an output by the MATLAB program GRNmap, you do not need to perform any further modifications to use the Workbook with GRNsight. You can move on to Section 2 to learn how to load your file as an input spreadsheet.
If you want to create manually a new file to use with GRNsight, format the file in the following manner:
-
GRNsight accepts Microsoft Excel Workbook (*.xlsx) files as its native format.
- Note that Excel 97-2003 Workbook (*.xls) files are not able to be read by GRNsight.
- If you have a SIF (*.sif) or GraphML (*.graphml) file, please see Section 1b-i.c or Section 1b-i.d, respectively, to verify that your files are formatted correctly for import into GRNsight.
-
GRNsight reads gene regulatory network information that is in the form of an adjacency matrix to generate the nodes and edges of the graph.
- There must be one worksheet in your Excel Workbook entitled “network” or “network_optimized_weights”. Generally, the “network” worksheet describes an unweighted network and the “network_optimized_weights” worksheet describes a weighted network. If the Workbook contains worksheets with both names, the display of the “network_optimized_weights” data will take precedence.
- In the “network” and “network_optimized_weights” worksheets, the upper-left cell (A1) should contain the text “cols regulators/rows targets”. This text is there as a reminder of the direction of the regulatory relationships specified by the adjacency matrix.
- The rest of the first row should contain the names of the transcription factors that are controlling the other genes in the network, one transcription factor name per column. Transcription factor names must be unique for each column and must be 12 characters or fewer.
- The rest of the first column should contain the names of the target genes that are being controlled by the transcription factors heading each of the columns in the matrix, one target gene name per row. Transcription factor names must be unique for each row and must be 12 characters or fewer.
- GRNsight is primarily designed to depict gene regulatory networks that contain just transcription factors and that can be represented by a symmetric adjacency matrix. However, starting with version 1.8, GRNsight allows the user to upload spreadsheets with asymmetric adjacency matrices. I.e., the number, identity, and order of genes across the top and side of the adjacency matrix do not have to match. A gene name at the top of the matrix will be considered the same as a gene name on the side if it contains the same text string, regardless of capitalization. For example, “ABF1”, “Abf1”, and “abf1” would all be considered the same gene, but “ABF_1” or “ABF-1” would not be considered the same as “ABF1”. Gene names may not contain any special characters other than "-" and "_". GRNsight is case-preserving in that it will keep the capitalization that the user originally entered into the spreadsheet. The gene name capitalization occurring across the top takes precedence over the gene name capitalization occurring on the side of the adjacency matrix for display of the gene names on the nodes of the graph. The nodes that display on the graph are automatically sized to accommodate the size of the label. Labels are restricted to 12 characters; use shorter gene symbols for nodes so that the node boxes do not get too large.
-
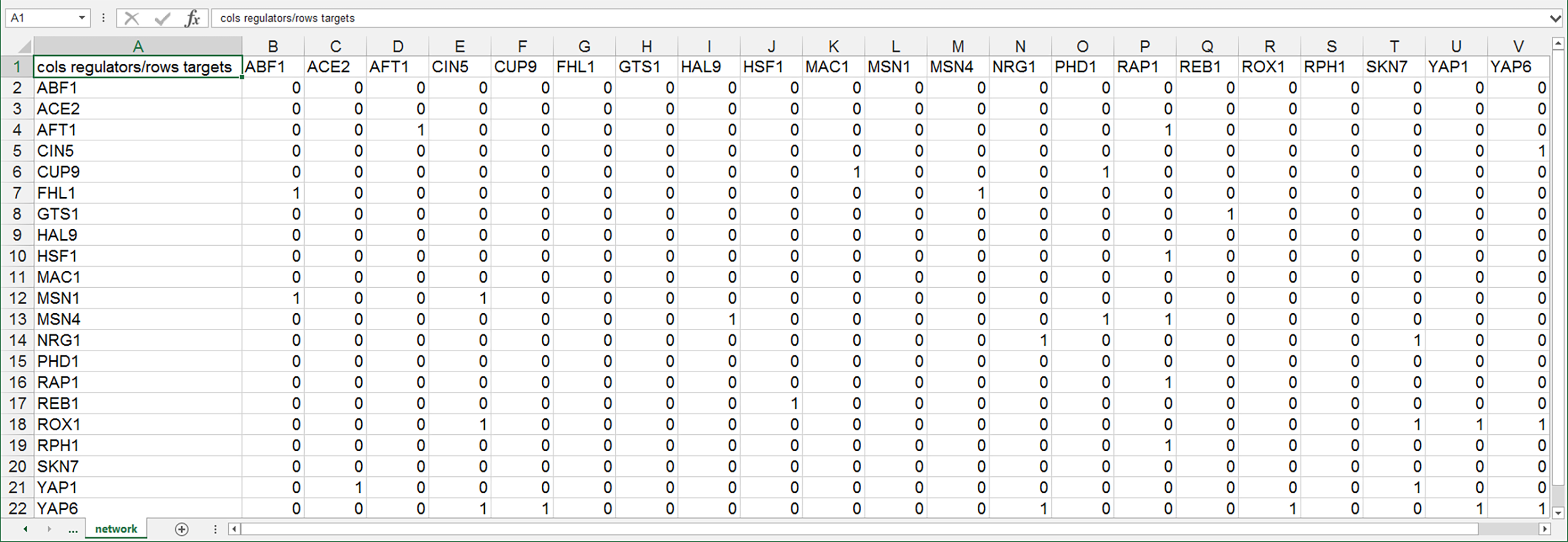
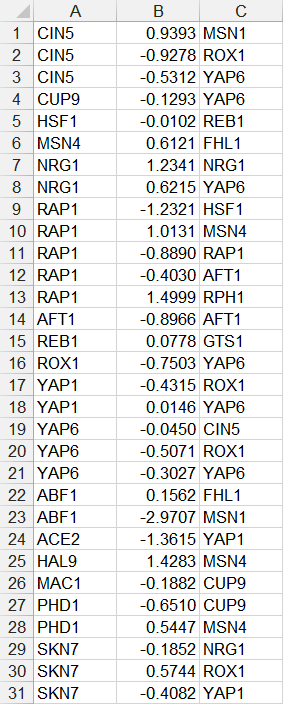
For unweighted networks in the “network” worksheet, each cell in the matrix should then contain a zero (0) if there is no regulatory relationship between those two transcription factors, or a one (1) if there is a regulatory relationship between them. In the screenshot below, the “1” in cell B7 means that ABF1 regulates FHl1. The “1” in cell D4 means that AFT1 regulates itself. When this file is loaded into GRNsight, it will generate a graph with 21 nodes, one for each transcription factor, and 31 edges, for each of the cells that contain a “1” in this adjacency matrix. The edges will be black with regular (pointy) arrowheads. The demonstration file "Demo #3: Unweighted GRN" corresponds to the adjacency matrix shown below. Follow the link to download the file.
-
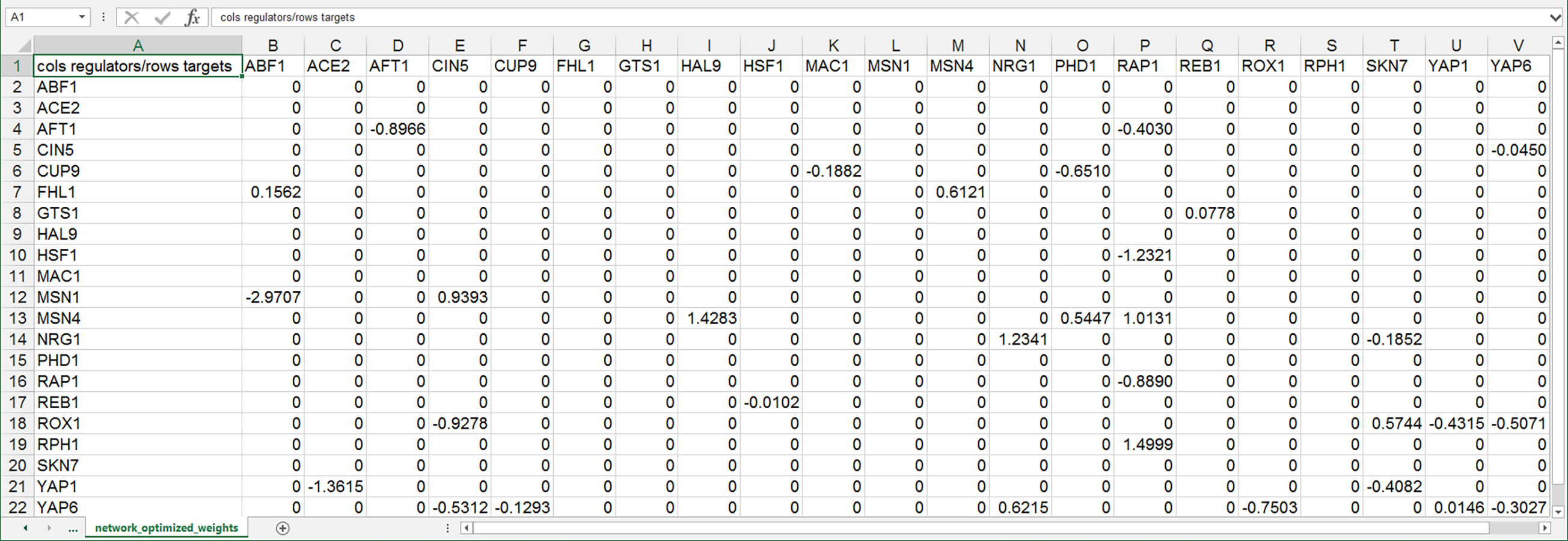
When GRNsight encounters a worksheet entitled “network” with all 1’s and 0’s specifying an adjacency matrix, it will automatically color the edges black and put regular (pointy) arrowheads on the edges. However, GRNsight can also be used to display weighted edges with varying thicknesses and colors based on the magnitude and sign of the weights, respectively. To generate this type of adjacency matrix by hand, create a worksheet called “network_optimized_weights”. Instead of using 1’s to indicate connections in the network, use values that are real numbers > 0 to indicate an activation relationship or < 0 to indicate a repression relationship. The screenshot below gives an example of this type of adjacency matrix. The demonstration file Demo #4: Weighted GRN corresponds to the adjacency matrix shown below. Follow the link to download the file. For the weighted edges to display with different colors and thicknesses, make sure that you have selected the menu item Edit > Preferences > Format edges based on optimized weight parameters. For more information on how GRNsight displays weighted edges on the graph, see Section 2 below.
- Note that GRNsight is designed to visualize “small-scale” or “medium-scale” gene regulatory networks, not the entire gene regulatory network for an organism. Currently, it is recommended that you upload networks with no more than 35 unique genes or 70 edges. If you upload a network with 50 nodes or 100 edges or more, you will receive a warning and the network graph will still display. If you attempt to upload a network of 75 or more nodes or 100 or more edges, the graph will not display.
- Follow these instructions to download and format a regulation matrix from the YEASTRACT database for use with GRNsight.
-
Starting in v3.0.0, GRNsight reads time series expression data in the form of an adjacency matrix, and displays this data as a node coloring visualization.
- Excel Workbook with names ending with "_expression" are processed as expression time series data. The upper-left cell (A1) should contain the text “id”.
- The rest of the first row should contain the names of the transcription factors under test, one transcription factor name per column. Transcription factor names must be unique for each column and must be 12 characters or fewer.
- The rest of the first column should contain numerical time points in ascending order.
- The values withing the adjancy matrix should represent the expression value of the particular transcription factor at a given time point. Blank cells are allowed, indicating the lack of a data point for a particular transcription factor at a given time.
- Please see Section 2e for more information on how to interpret and manipulate the node coloring visualization.
-
Starting in v4.0.0 GRNsight reads metadata from a sheet entitled “optimization_parameters.”
- The header should contain “optimization” cell A1 and “value” in cell B1.
- The following optimization parameters can be a number: “alpha”, “kk_max”, “MaxIter”, “TolFun”, “MaxFunEval”, “TolX”, “L_curve”, “estimate_params”, “make_graphs”, “fix_P”, “fix_b”, “taxon_id”, and “b_or_tau”.
- The following optimization parameters can be a string: “production_function” and “species”.
- The following optimization parameters can be a list of numbers: “expression_timepoints” and “simulation_timepoints”.
- The following optimization parameters can be a list of numbers: “expression_timepoints” and “simulation_timepoints”.
- Navigate to Generate Regulation Matrix on the YEASTRACT site.
- Select the appropriate radio buttons/check boxes for the filters.
- Paste a list of transcription factors into the appropriate field.
- Paste a list of targets into the Target ORF/Genes field, or check the box to consider all ORF/Genes.
- Click the Generate button.
- In the results window that appears, click the link to download the Regulation matrix results file as a Semicolon Separated Values (CSV) file (not the Two-column table, Tab Separated Values (TSV) file).
- Once you have downloaded the file, launch Microsoft Excel.
- Select the menu item, File > Open and select the file that you downloaded.
- Select Column A.
- Select the menu item, Data > Text to Columns...
- In the first window of the wizard that appears, select the radio button for "Delimited" and click Next.
- In the second window of the wizard that appears, check the box for "Other" under "Delimiters" and type a semicolon in the field to the right and click Finish.
- Select the menu item, File > Save As... and save the file as an Excel Workbook (.xlsx).
- YEASTRACT organizes the adjacency matrix with the transcription factors as rows and the target genes as columns, which is opposite to the format that GRNmap and GRNsight expects. To flip this orientation, first create a new worksheet by clicking on the new worksheet icon at the bottom of the screen. Name this new worksheet "network".
- Select the adjacency matrix from the first worksheet and copy it to the clipboard. Go to the "network" worksheet and click on cell A1. Select the menu item Edit > Paste Special. In the window that appears, check the box "Transpose" and click OK.
- The labels for the genes in the columns and rows needs to match. Thus, delete the "p" from each of the gene names in the columns.
- Paste the following text into cell A1 "cols regulators/rows targets".
- Save your work. This file is now ready for loading into GRNsight. You can choose to delete the original worksheet or keep it in your workbook.
- A SIF file is a tab-delimited text file with the file extension .sif.
- Note that the Cytoscape SIF specification also allows space-delimited files. However, due to the confusion caused when a space character is used in a node label or interaction type, GRNsight only allows tabs as delimiters for .sif files.
- Lines in the SIF file specify a source node, a relationship type (or edge type), and one or more target nodes separated by tab characters.
- For a network defined in SIF format, node names should be unique, as identically named nodes will be treated as identical nodes.
- Node names must be 12 characters or fewer.
- Starting from GRNsight v2, the relationship type for a weighted network is required to be "pd". "pd" was chosen because it commonly means protein -> DNA (e.g. a regulatory transcription factor binding upstream of a target gene) in the systems biology community. A strict requirement for the relationship type was specified to expand upon SIF syntactic error checking capabilities.
- Duplicate entries are ignored (as would occur if the same source-target pair was listed twice with two different relationship types).
- GRNsight automatically detects whether the SIF file represents an unweighted or weighted network through the examination of the relationship type. The screenshots and descriptions below are four valid ways that an unweighted network can be specified as a SIF file, using the file Demo #3: Unweighted GRN" as an example. (The screenshots display a tab-delimited text file as it would be displayed in Microsoft Excel.) Follow the link to download a sample SIF file.
- In the first example below, each source node in the network is listed in the first column, the relationship type “pd” is listed in the second column, and the target node is listed in the third column. If a node has multiple target nodes, it is listed multiple times, one relationship per line. A node may regulate itself (for example, AFT1). Note that FHL1, GTS1, MSN1, and RPH1 do not have any target nodes, but are still listed as source nodes. While they can be listed, it is not necessary to do so, as is shown in the second example.
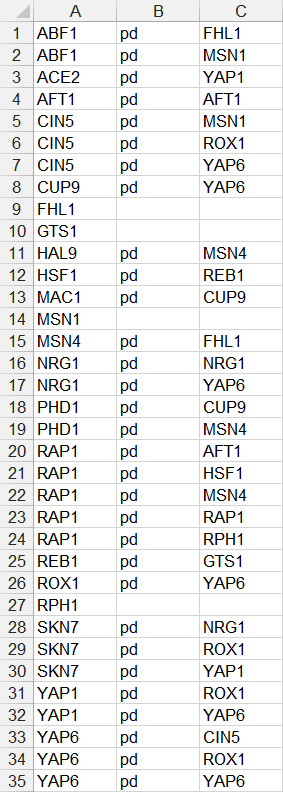
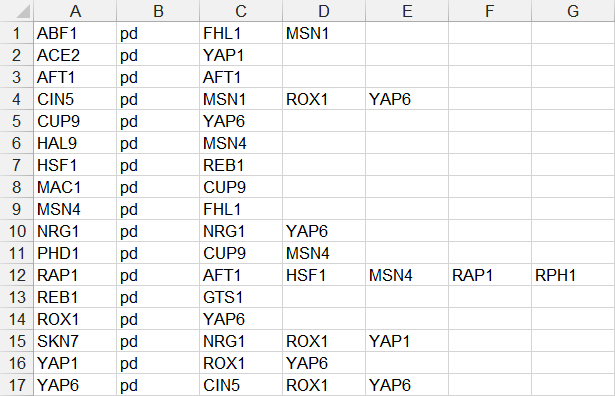
- In the second example below, only source nodes that have target nodes are listed in the first column, the relationship type “pd” is listed in the second column, and the target node is listed in the third column. If a node has multiple target nodes, it is listed multiple times, one relationship per line. (FHL1, GTS1, MSN1, and RPH1 were removed because they did not have any target nodes.)
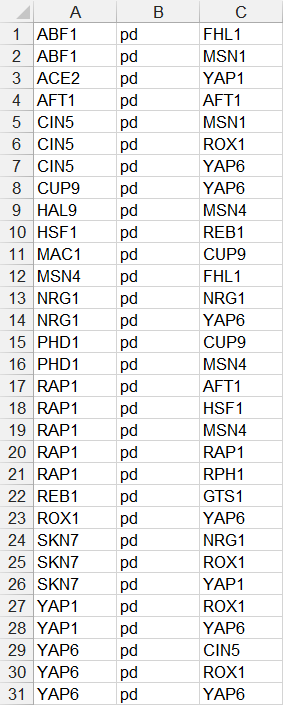
- In the third example below, each source node is only listed once on one line, followed by the relationship type “pd”, followed by a series of target nodes, separated by tab characters. Note that FHL1, GTS1, MSN1, and RPH1 do not have any target nodes, but are still listed as source nodes. While they can be listed, it is not necessary to do so, as is shown in the fourth example.
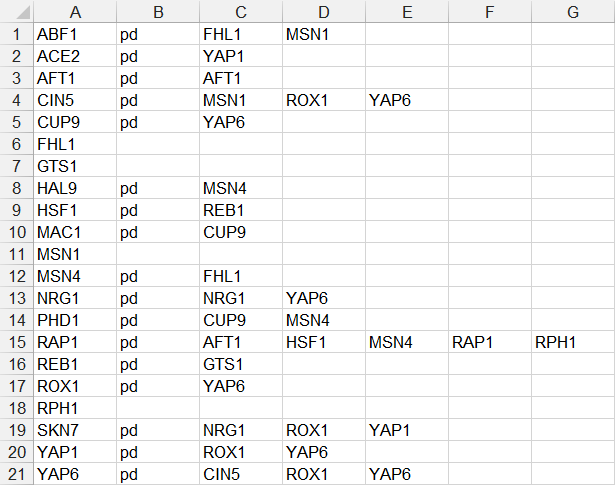
- In the fourth example below, each source node is only listed on one line, followed by the relationship type “pd”, followed by a series of target nodes, separated by tab characters. (FHL1, GTS1, MSN1, and RPH1 were removed because they did not have any target nodes.)
- A Cytoscape SIF file is only intended to convey the network nodes and edge connections; it does not support the storage of other node or edge attributes or display properties. However, with a modification to the relationship type, a SIF file can represent a weighted network and be displayed as such in GRNsight upon import (See Section 2a).
- To encode a weighted network in a SIF file, instead of using a text string like “pd” as the relationship type, substitute the numerical weight value. Use real numbers > 0 to indicate an activation relationship or < 0 to indicate a repression relationship. The screenshots and descriptions below are two valid ways that a weighted network can be specified as a SIF file, using the file Demo #4: Weighted GRN" as an example. (The screenshots display a tab-delimited text file as it would be displayed in Microsoft Excel.) Follow the link to download a sample SIF file.
- In the first example below, each source node in the network is listed in the first column, the numerical value of the weight is listed in the second column, and the target node is listed in the third column. If a node has multiple target nodes, it is listed multiple times, one relationship per line. Note that FHL1, GTS1, MSN1, and RPH1 do not have any target nodes, but are still listed as source nodes. While they can be listed, it is not necessary to do so, as is shown in the second example.
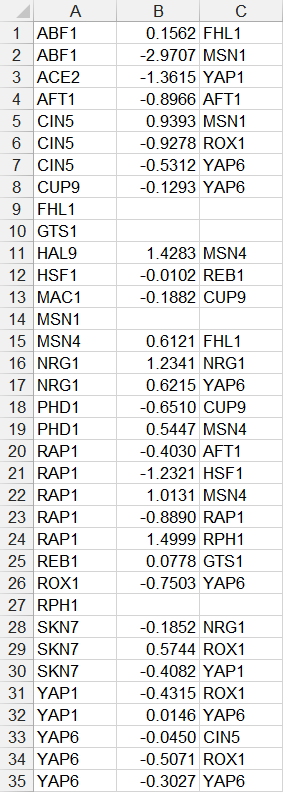
- In the second example below, only source nodes that have target nodes are listed in the first column, the numerical value of the weight is listed in the second column, and the target node is listed in the third column. If a node has multiple target nodes, it is listed multiple times, one relationship per line. (FHL1, GTS1, MSN1, and RPH1 were removed because they did not have any target nodes.)
- For a weighted network, each source-to-target relationship should be on a separate line. While it is possible that two or more source-to-target relationships would have the same exact weight value, it is not recommended to use the shorthand of listing multiple target nodes on the same row.
- GRNsight will automatically detect a SIF file representing a weighted network when it finds numerical values as the relationship type. It will change the arrowhead type and the thickness and color of the edges as described in Section 2a.
- However, if it detects a mix of numerical and "pd" string values as the relationship type, it will issue a warning to the user that this was detected, but still display the network as an unweighted network.
- If the user has selected the menu item Edit > Preferences > Default to black edges with regular arrowheads, GRNsight will ignore the numerical weight values and display the network as an unweighted graph.
- Note that these “weighted” SIF files are able to be imported in to Cytoscape (tested with v3.4.0), but the weight values will not be properly stored as edge attributes by Cytoscape because it does not use the SIF format in this way.
- Note that GRNsight is designed to visualize “small-scale” or “medium-scale” gene regulatory networks, not the entire gene regulatory network for an organism. Currently, it is recommended that you import networks with no more than 35 unique genes or 70 edges. If you upload a network with 50 nodes or 100 edges or more, you will receive a warning and the network graph will still display. If you attempt to upload a network of 75 or more nodes or 100 or more edges, the graph will not display.
Users who have data in GraphML format can import these files directly into GRNsight. However, if you are creating a new input file for use with GRNsight, we recommend using the native Excel format expected by GRNsight as described in Section 1a.
A primer for the GraphML format can be found at http://graphml.graphdrawing.org/primer/graphml-primer.html, whereas the detailed specification can be found at http://graphml.graphdrawing.org/specification.html. The instructions below borrow heavily from the GraphML primer.
- A GraphML file is an Extensible Markup Language (XML) file with the extension .graphml.
- GraphML has the ability to specify graph features that GRNsight cannot display. In those cases, GRNsight will ignore the features that it does not support.
- The minimum requirements for an unweighted GraphML file that GRNsight can read and display is best illustrated by an example. The screenshots below show the top and bottom of the Demo #3 Unweighted GRN (21 genes, 31 edges) GraphML file as exported by GRNsight. (The screenshots were created from a view of this file in the firstobject XML Editor). Click the link to download the file.

...

- “The first line of the document is an XML process instruction which defines that the document adheres to the XML 1.0 standard and that the encoding of the document is UTF-8, the standard encoding for XML documents.”—GraphML Primer
- “The second line contains the root-element element of a GraphML document: the
graphmlelement. The graphml element, like all other GraphML elements, belongs to the namespacehttp://graphml.graphdrawing.org/xmlns. For this reason we define this namespace as the default namespace in the document by adding the XML Attributexmlns="http://graphml.graphdrawing.org/xmlns"to it. The two other XML Attributes are needed to specify the XML Schema for this document. In our example we use the standard schema for GraphML documents located on thegraphdrawing.orgserver. The first attribute,xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance", definesxsias the XML Schema namespace. The second attribute,xsi:schemaLocation="http://graphml.graphdrawing.org/xmlns http://graphml.graphdrawing.org/xmlns/1.0/graphml.xsd", defines the XML Schema location for all elements in the GraphML namespace.”—GraphML Primer - The third line is a comment injected by GRNsight, giving the name of the program that created the document (GRNsight), the version (in this case v1.18), URL of the GRNsight home page, and URL to the GitHub release page for the specified version. This information is provided upon export of a GraphML file by GRNsight to assist with keeping track of the provenance of the data, and is optional for the import of GraphML files into GRNsight.
- The next three lines are key elements. GraphML standardizes only the representation of nodes and edges and their directions; all other characteristics, such as names, weights, and other values, are left for others to specify through a key element, which is not subject to a controlled vocabulary. For an unweighted network, GRNsight-exported GraphML defines the keys:
- name for the node label (The node name is defined for compatibility with Cytoscape, but is optional for GRNsight to read and display the graph.)
- interaction for the relationship type of the edge (The relationship type is defined in the same way as for SIF files described in Section 1c for compatibility with Cytoscape, but is optional for GRNsight to read and display the graph.)
- name for the edge (The edge name is defined for compatibility with Cytoscape, but is optional for GRNsight to read and display the graph.)
- "A graph is, not surprisingly, denoted by a
graphelement. Nested inside agraphelement are the declarations of nodes and edges. A node is declared with anodeelement, and anedgewith an edge element."—GraphML Primer - In the graph element in line 7, the edgedefault attribute is set to “directed”. GRNsight will only display directed graphs, not undirected or mixed graphs.
- The id attribute gives the name of the GraphML file, and is optional for GRNsight to read and display the graph. The id is provided for compatibility with Cytoscape.
- The next three elements in the screenshot are node declarations, giving an id and name for the nodes “ABF1”, “ACE2”, and “AFT1”.
- Note that for GRNsight-exported GraphML, the node id and name are identical and unique to the file. However, other programs may have a node name that is different than the node id. For example, both Cytoscape v3.4.0 and yED v3.16 assign internal node identifiers that the user cannot edit. The user does have the ability edit the node label, and it is for this reason that GRNsight will preferentially display the node label on the graph. However, user editability in Cytoscape and yED leads to a situation where a user can actually assign the same label to two different nodes. In this case, duplicate node labels will be displayed by GRNsight, and the user is directed to go back and edit the source file.
- Cytoscape uses the name key to assign a label to nodes and a shared name key to assign a label to the root network when subnetworks are present.
- yED uses a y:NodeLabel key to assign the label to nodes.
- In order to maximize the ability for GRNsight to read GraphML exported from Cytoscape and yED, the following conditions are followed when displaying a graph: if all three keys are present name gets top priority, followed by shared name, then y:NodeLabel, then id.
- The next three elements shown in the screenshot are edge declarations for the edges ROX1-to-YAP6, YAP1-to-YAP6, and YAP6-to-YAP6.
- The edge element attributes source and target give the node id for the source and target node, respectively.
- The interaction key is the relationship type, similar to what is defined for SIF files. It is given for compatibility with Cytoscape and defaults to “pd” for protein->DNA, but is otherwise ignored by GRNsight for display.
- The edge name key is given for compatibility with Cytoscape, but is otherwise ignored by GRNsight for display.
- As noted above, GraphML has the ability to specify graph features that GRNsight cannot display. In those cases, GRNsight will ignore these features. The original imported file will not be changed, but a GRNsight re-export of the same graph will omit these elements.
- GRNsight cannot display a nested graph. Any links that connect nodes from the main graph to nodes in the subgraph will not display.
- Any other specifications for the display of the graph (layout, colors, etc.) will be ignored by GRNsight.
- We have tested the display of networks exported in GraphML format from Cytoscape v3.4.0 and yED v3.16, but because the flexibility of the GraphML standard enables divergence in implementation, we cannot guarantee that GraphML exported from other programs will be read and displayed correctly by GRNsight. If you encounter problems, please submit an issue to GRNsight @ GitHub (requires free GitHub account).
- The minimum requirements for a weighted GraphML file that GRNsight can read is best illustrated by an example. The screenshots below show the top and bottom of the Demo #4 Weighted GRN (21 genes, 31 edges, Schade et al. 2004 data) GraphML file as exported by GRNsight. Click the link to download the file.

...

- Note that there are only two major differences between the unweighted and weighted GraphML formats that GRNsight exports:
- A key declaration defining a weight attribute for an edge as the datatype “double”.
- Note that there is currently a bug in Cytoscape v3.4.0 with the way it exports GraphML files. It currently assigns the weight attribute for a node instead of an edge. To maximize the ability for GRNsight to read and display Cytoscape-exported GraphML, GRNsight does not currently enforce that the weight attribute be declared "for an edge". The bug has been reported to the Cytoscape group.
- The addition of the weight key for each edge. For example, the weight for the edge ROX1-to-YAP6 is -0.7503313690638223.
- GRNsight automatically detects if a GraphML file specifying a weighted graph has been imported.
- For a weighted graph to display, the weight attribute must be present and valid for all edges declared in the graph. If one or more weights are missing, GRNsight will display the graph as unweighted with black edges and pointed arrowheads.
- Note that GRNsight is designed to visualize “small-scale” or “medium-scale” gene regulatory networks, not the entire gene regulatory network for an organism. Currently, it is recommended that you import networks with no more than 35 unique genes or 70 edges. If you upload a network with 50 nodes or 100 edges or more, you will receive a warning and the network graph will still display. If you attempt to upload a network of 75 or more nodes or 100 or more edges, the graph will not display.
- Once you have your appropriately formatted Excel Workbook file (*.xlsx), you can load it into GRNsight.
- Go to the GRNsight home page. Select the menu option File > Open. A dialog box will open for you to select your file. Clicking the Open button will load your file. Clicking the Cancel button will return you to the GRNsight home page.
- The name of the file that is currently loaded will appear in the menu bar. The number of nodes and edges in the graph will display on the right-hand side of the menu bar.
- To load a different file, select the menu option File > Open to load a GRNsight-formatted .xlsx file or File > Import to import a file in SIF or GraphML.
- To reload the same file, select the menu option File > Reload. Note that the Reload menu item will be disabled until a file has been loaded.
- If you do not have your own input spreadsheet, you can load and view demonstration gene regulatory networks. Selecting the menu item Demo > Demo #1: Unweighted GRN (15 genes, 28 edges, Dahlquist Lab unpublished data) or Demo #3: Unweighted GRN (21 genes, 31 edges) will load a network graph with black edges and regular (pointy) arrowheads. Selecting the menu item Demo > Demo #2: Weighted GRN (15 genes, 28 edges, Dahlquist Lab unpublished data) or Demo #4: Weighted GRN (21 genes, 31 edges, Schade et al. 2004 data) will load a network graph with colored edges of varying thicknesses. Demos #3 and 4 are based on modeling reported in the following publication: Dahlquist, K.D., Fitzpatrick, B.G., Camacho, E.T., Entzminger, S.D., and Wanner, N.C. (2015) Parameter Estimation for Gene Regulatory Networks from Microarray Data: Cold Shock Response in Saccharomyces cerevisiae. Bulletin of Mathematical Biology. 77: 1457-1492, published online 29 September 2015, DOI: 10.1007/s11538-015-0092-6, which, in turn, is based on microarray data from this publication: Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY (2004) Cold adaptation in budding yeast. Mol Biol Cell 15:5492–5502. doi:10.1091/mbc.E04-03-0167. See Section 2 for details on how GRNsight displays the graph.
- Once you have your appropriately formatted SIF file (*.sif; see Section 1a-ii.c), you can import it into GRNsight.
- Go to the GRNsight beta page. Select the menu option File > Import > SIF…. A dialog box will open for you to select your file. Clicking the Open button will load your file. Clicking the Cancel button will return you to the GRNsight home page.
- The name of the file that is currently loaded will appear in the menu bar. The number of nodes and edges in the graph will display on the right-hand side of the menu bar.
- To reload the same file, select the menu option File > Reload. Note that the Reload menu item will be disabled until a file has been imported.
- To load a different file, select the menu option File > Open to load a GRNsight-formatted .xlsx file or File > Import to import a file in SIF or GraphML format.
- GRNsight will automatically detect a SIF file representing a weighted network when it finds numerical values as the relationship type. It will change the arrowhead type and the thickness and color of the edges as described in Section 2a.
- However, if GRNsight detects a mix of numerical and text string values or missing values for the relationship type, it will display the network as an unweighted network.
- If the user has selected the menu item Edit > Preferences > Default to black edges with regular arrowheads, GRNsight will ignore the numerical weight values and display the network as an unweighted graph.
- Once you have your appropriately formatted GraphML file (*.graphml), you can import it into GRNsight.
- Go to the GRNsight beta page. Select the menu option File > Import > GraphML…. A dialog box will open for you to select your file. Clicking the Open button will load your file. Clicking the Cancel button will return you to the GRNsight home page.
- The name of the file that is currently loaded will appear in the menu bar. The number of nodes and edges in the graph will display on the right-hand side of the menu bar.
- To reload the same file, select the menu option File > Reload. Note that the Reload menu item will be disabled until a file has been imported.
- To load a different file, select the menu option File > Open to load a GRNsight-formatted .xlsx file or File > Import to import a file in SIF or GraphML format.
- GRNsight will automatically detect a GraphML file representing a weighted network when it finds numerical values stored as weight attributes for the edges. It will change the arrowhead type and the thickness and color of the edges as described in Section 2a.
- However, if GRNsight detects a mix of numerical and text string values or missing values for the weight attributes, it will display the network as an unweighted network.
- If the user has selected the menu item Edit > Preferences > Default to black edges with regular arrowheads, GRNsight will ignore the numerical weight values and display the network as an unweighted graph.
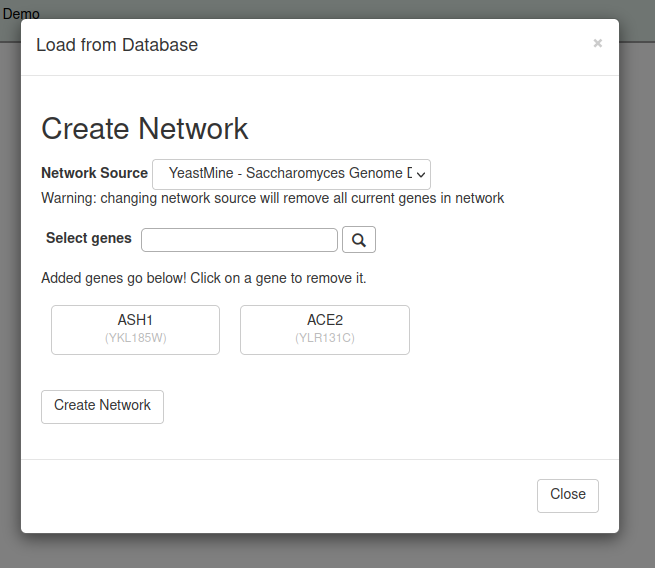
If you do not have your own input spreadsheet, you have the option to select the specified genes in your gene regulatory network using the Load from Network feature.
-
To access the Load from Database feature Select the side bar or top menu item Network > Load From Database. From there a pop up window should appear where you can choose which network source you want to select your genes from, and search for which genes you would like to add to your network. You can search using the standard-like name or systematic-like name (in referece to the standard and systematic names of Saccharomyces cerevisiae). Once selected the genes will appear listed below the search bar. To remove a gene, click on the selected gene and it will be removed from the network. Once complete, click on the Create Network button and the selected gene regulatory network will load into GRNsight.
GRNsight graphs are generated through D3.js, a JavaScript data visualization library. We have modified the default implementation of D3 to include the following features:
- Nodes are rectangular.
- The labels on the nodes are what is found in your “network” or “network_optimized_weights” worksheet, depending on whether you are displaying an unweighted or weighted graph. GRNsight is case-preserving in that it will keep the capitalization that the user originally entered into the spreadsheet. The gene name capitalization occurring across the top takes precedence over the gene name capitalization occurring on the side of the adjacency matrix for display of the gene names on the nodes of the graph.
- The nodes are automatically sized to accommodate the size of the label. Labels are restricted to 12 characters; use shorter gene symbols for nodes so that the node boxes do not get too large.
- Edges are arcs, but when two nodes get close together, the edges become straight lines. Self-regulatory edges are indicated by a loop on the lower-right side of a node.
GRNsight has two layouts, grid layout and force graph layout. Grid layout sorts the nodes in alphabetical order and organizes them in a grid pattern. This ultimately gives the user power to compare graphs in easier, faster, and more efficient way.
Note that GRNsight is designed to visualize “small-scale” or “medium-scale” gene regulatory networks, not the entire gene regulatory network for an organism. Currently, it is recommended that you upload networks with no more than 35 unique genes or 70 edges. If you upload a network with 50 nodes or 100 edges or more, you will receive a warning and the network graph will still display, although it may just look like “spaghetti”. If you attempt to upload a network of 75 or more nodes or 100 or more edges, the graph will not display.
If your adjacency matrix is contained in a worksheet entitled “network” and contains just 1's and 0's, GRNsight will make the edges connecting nodes black with a regular (pointy) arrowhead originating from the transcription factor (columns) to the target gene (rows) in your matrix. (Note that selecting the menu item Edit > Preferences > Default to black edges with regular arrowheads will make the graph display as an unweighted graph even if the worksheet "network_optimized_weights" has real numbers instead of 1's to indicated weighted edges.)
If your adjacency matrix is contained in a worksheet entitled “network_optimized_weights” and contains real numbers instead of just 1’s to indicate weighted connections, it will adjust the arrowhead type, thickness, and color of the edges as follows:
- For positive weights > 0, the edge will be given a regular (pointy) arrowhead to indicate an activation relationship between the two nodes.
- For negative weights < 0, the edge will be given a blunt arrowhead (a line segment perpendicular to the edge direction) to indicate a repression relationship between the two nodes.
- The thickness of the edge will vary based on the magnitude of the absolute value of the weight. Larger magnitudes will have thicker edges and smaller magnitudes will have thinner edges. The way that GRNsight determines the edge thickness is as follows. GRNsight divides all weight values by the absolute value of the maximum weight in the matrix to normalize all the values to between zero and 1. GRNsight then adjusts the thickness of the lines to vary continuously from the minimum thickness (for normalized weights near zero) to maximum thickness (normalized weights of 1). Normalization can be set using the box in the sidebar that appears once weighted graphs are loaded. Once set, the normalization can also be reset to the defaul value.
- The color of the edge also imparts information about the regulatory relationship. Edges with positive normalized weight values from 0.05 to 1 are colored red (magenta in GRNsight < v3.0.0); edges with negative normalized weight values from -0.05 to -1 are colored blue (cyan in GRNsight < v3.0.0). Edges with normalized weight values between -0.05 and 0.05 are colored gray to emphasize that their normalized magnitude is near zero and that they have a weak influence on the target gene. The threshold to show gray weights can be manipulated by using the Gray Threshold Slider. The threshold is based on 1% increments of the maximum weight. Optionally, users may specify that a gray edge be displayed as dashed, to provide additional visual indication of weak relationships.
- For the weighted edges to display with different colors and thickness, make sure that you have selected the menu item Edit > Preferences > Format edges based on optimized weight parameters. Selecting the menu item Edit > Preferences > Default to black edges with regular arrowheads will make the graph display as an unweighted graph even if the worksheet "network_optimized_weights" has real numbers instead of 1's to indicated weighted edges.
- If you hover your mouse over an edge on the graph, a tooltip will display the weight value to four significant digits. This behavior can be changed by selecting options under the "Hide/Show Edge Weights" sidebar menu header. "Always Show Edge Weights" will cause edge weights to display even when not moused over. "Never Show Edge Weights" will prevent any edge weights from showing up even if an edge is moused over.
Once you have selected the spreadsheet file to be loaded, GRNsight automatically lays out the positions of the nodes and edges based on certain Force Graph Parameters.
- Link Distance determines the minimum distance maintained between nodes, i.e., it changes the length of the edges. Smaller numbers specify shorter lengths, larger numbers specify longer lengths. The range of values for this parameter are 1 to 1000 with a default value of 500.
- Charge changes the strength of the force that causes the nodes to repel each other. A more negative value causes more repulsion between nodes. The range of values for this parameter are -2000 to 0 with a default value of -1000.
- You may change the values of each of the force graph parameters by moving the sliders back and forth.
- Checking the box next to “Lock Force Graph Parameters” locks the current settings of the parameters (positions of the sliders) in place so that they cannot be changed. Unchecking the box unlocks the sliders again. Alternately, you can select the menu item Format > Lock Force Graph Parameters to lock the current settings. A check mark will appear next to the menu item to indicate that they are locked. Selecting the menu item Format > Lock Force Graph Parameters a second time will unlock the sliders again.
- Clicking the “Reset Force Graph Parameters” button will restore the default settings. Alternately, you can select the menu item Format > Reset Force Graph Parameters to restore the default settings.
- Clicking the “Undo Reset” button will restore the Force Graph Parameter settings that were in place before the Force Graph Parameters were reset either by clicking the button or selecting the menu item. “Undo Reset” is also a menu item under the Format menu.
- The force graph parameter sliders, lock force graph parameter sliders check box and menu item, and reset force graph parameter button and menu item are all active before a spreadsheet is loaded. The values set before loading will be used when a spreadsheet is loaded.
- Note that reloading the input file with the same Force Graph Parameter settings (even the defaults) will not necessarily lead to the exact same layout of the graph due to some randomness in D3’s layout algorithm.
The bounding box can be changed between three different sizes, or fit to the available space in the window. The initial default size that best fits the available space on the screen is selected when GRNsight is loaded, though this setting can be changed at any time. As the network loads, it will by default expand the bounding box outwards until the force graph parameters have fully been applied. The full size of the expanded graph can be seen using zooming or scrolling. The graph can be zoomed using the mouse wheel, as well as the on-screen zoom slider. The graph can be scrolled by clicking and draggong, or by using the arrow buttons. Should the graph go off-screen, the sun button in the center of the arrow buttons will center the graph. Choosing the "Restrict Graph to Viewport" option will prevent this expansion, and revert GRNsight to the behavior seen in versions before v2.0.20. The available space in the bounding box on restricted graphs will vary based on the size of the viewport, and will be indicated by a light gray border that can be seen upon zooming out. Note that if the graph is being restricted to the viewport, it will not be able to be scrolled past the visible area of the viewport. It also will not be able to be zoomed in past the bounding box.
By default, when a user uploads an Excel workbook which contains correctly formatted expression sheets, nodes are colored with a heat map overlay to visualize time series expression data. Expression values of a transcription factor at a given time point are mapped to corresponding vertical slices on the node, and colored according to the node coloring legend located in the sidebar. Blue slices represent negative expression values (indicating a decrease in mRNA), while red slices represent positive expression values (indicating an increase in mRNA). The greater the magnitude of the expression value, the darker the shade of red or blue.
Users may adjust the following node coloring parameters, located in both the sidebar and dropdown menus:
-
Node Coloring Toggle Button
Turns node coloring on or off. -
Top Dataset
Users may select the dataset used to generate the top half of the node coloring visualization. Options in the dropdown list are automatically populated with names of detected expression sheets. In GRNsight v5.0.0, users may also select an expression sheet from our Expression Database. -
Average Replicate Values (Top Dataset)
If this option is checked, GRNsight will average replicate expression values taken at the same timepoint for the top half of node coloring. -
Bottom Dataset
Users may select the dataset used to generate the bottom half of the node coloring visualization. Options in the dropdown list are automatically populated with names of detected expression sheets. In GRNsight v5.0.0, users may also select an expression sheet from our Expression Database. -
Average Replicate Values (Bottom Dataset)
If this option is checked, GRNsight will average replicate expression values taken at the same timepoint for the bottom half of node coloring. -
Log Fold Change Max Value
Indicates the value that will map to the darkest red/blue, and changes the node coloring legend accordingly. A negative log fold change max value will flip the red/blue scale. The default log fold change max value is 3. -
Node Coloring for PPI
If a PPI network file is loaded and includes valid gene names, node coloring can be toggled and color the nodes according to the expression data. Note: We are overlaying mRNA-level data onto a PPI network.
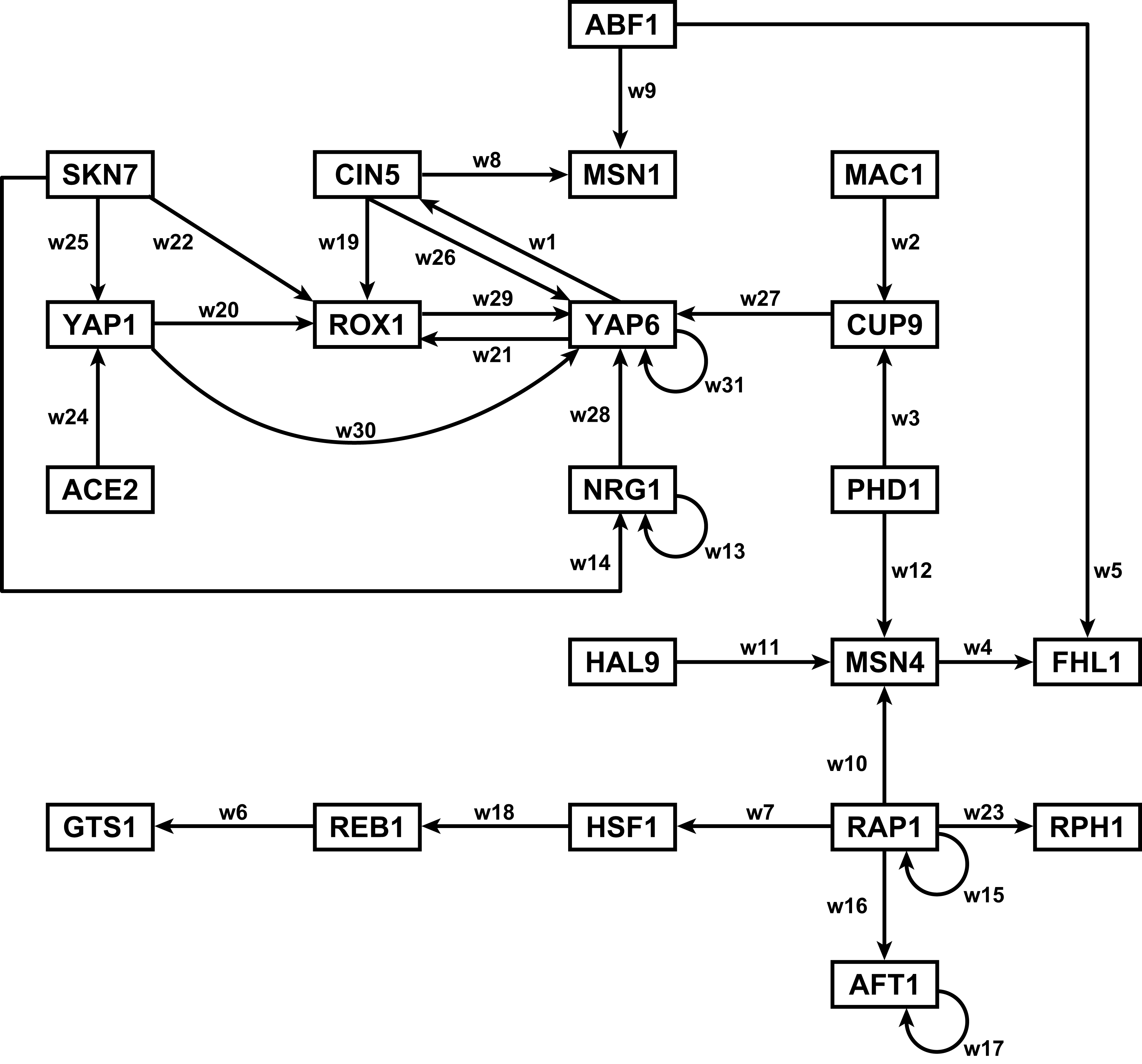
Section 2 above described how GRNsight uses the data in the adjacency matrix to automatically display a graph. This section describes how to interpret the results of the resulting gene regulatory network graph biologically, focusing on the demonstration files Demo #3: Unweighted GRN (21 genes, 31 edges) and Demo #4: Weighted GRN (21 genes, 31 edges, Schade et al. 2004 data). These two files describe gene regulatory networks from budding yeast, Saccharomyces cerevisiae, and correspond to supplementary data published in the paper, Dahlquist, K.D., Fitzpatrick, B.G., Camacho, E.T., Entzminger, S.D., and Wanner, N.C. (2015) Parameter Estimation for Gene Regulatory Networks from Microarray Data: Cold Shock Response in Saccharomyces cerevisiae. Bulletin of Mathematical Biology. 77: 1457-1492, published online 29 September 2015, DOI: 10.1007/s11538-015-0092-6, and when displayed by GRNsight, represent interactive versions of Figures 1 and 8 of that paper respectively.
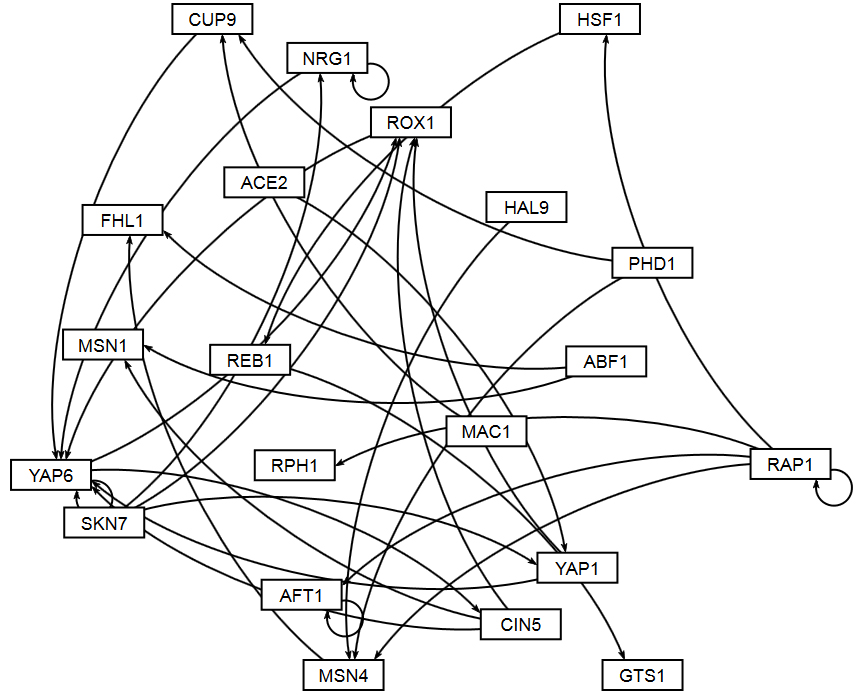
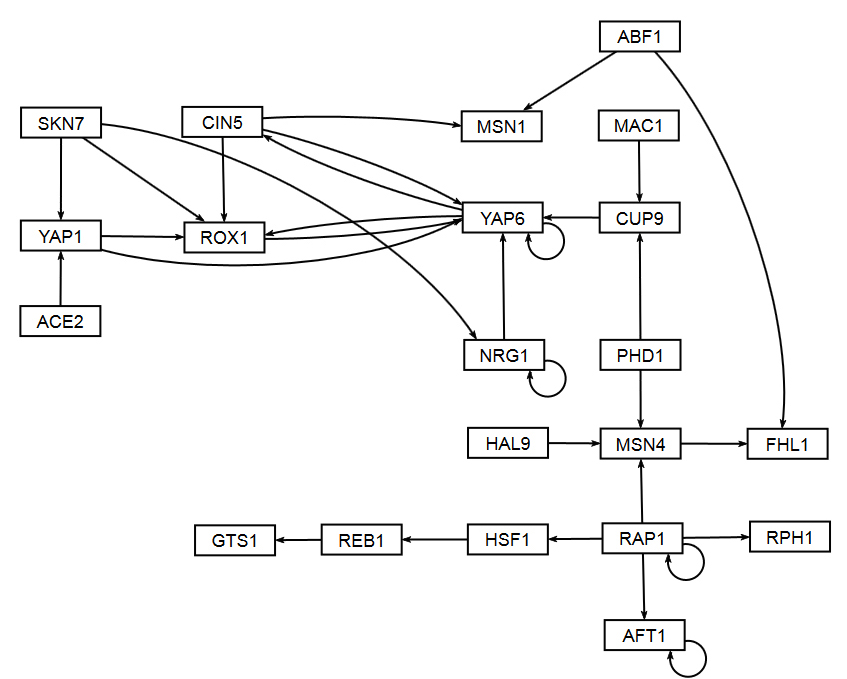
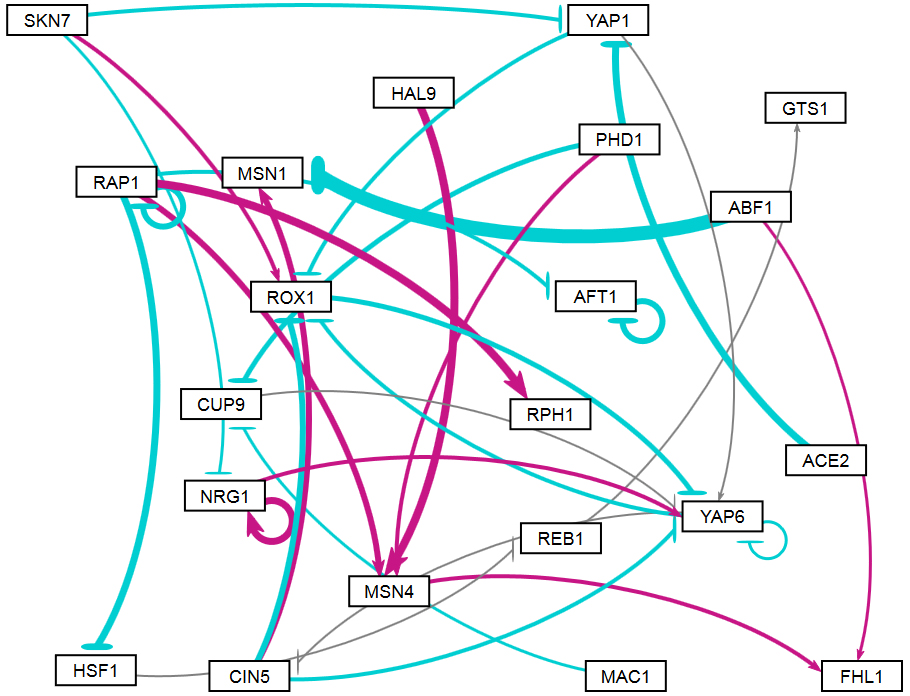
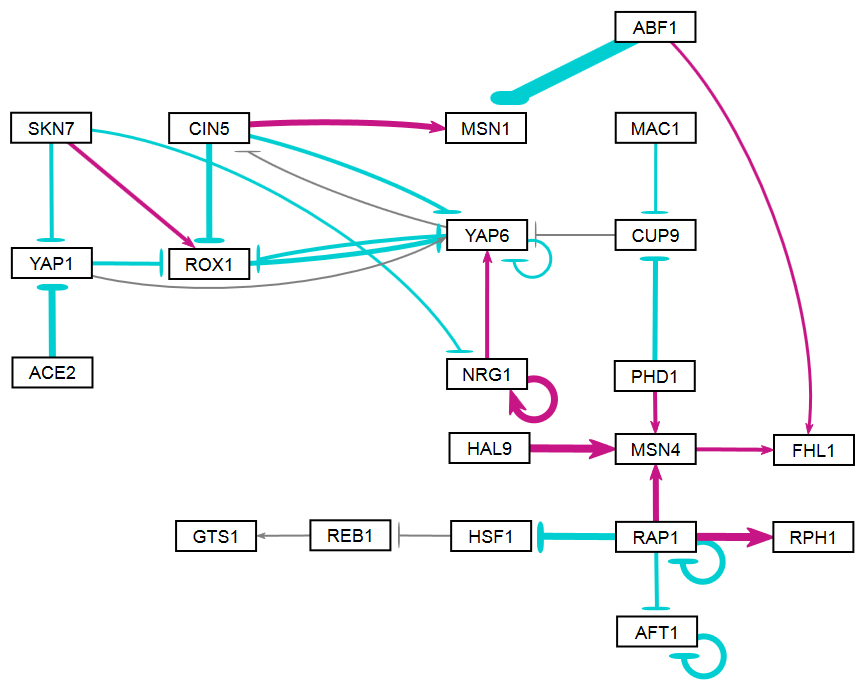
The figures shown below give a side-by-side view of the same adjacency matrices laid out by GRNsight and by hand. The top three figures are derived from Demo #3: Unweighted GRN (21 genes, 31 edges), and the bottom three panels are derived from Demo #4: Weighted GRN (21 genes, 31 edges, Schade et al. 2004 data). The left two panels show an example of the automatic layout performed by GRNsight. The right two panels show the same adjacency matrix laid out by hand in Adobe Illustrator, corresponding to Figure 1 (top right) and Figure 8 (bottom right) of the Dahlquist et al. (2015) paper. The middle two panels started with the automatic layout from GRNsight and then were manually manipulated from within GRNsight to lay them out similarly to the right-hand panels. The use of GRNsight represents a substantial time savings compared to creating the same figure entirely by hand.
 |
 |
 |
 |
 |
 |
- Viewing the unweighted network (top panel) allows one to make observations about the network structure. For example, YAP6 has the highest in-degree, being regulated by six other transcription factors. RAP1 has an out-degree of five, regulating four other transcription factors and itself. Four genes, AFT1, NRG1, RAP1, and YAP6, regulate themselves. Many of the transcription factors are involved in regulatory chains, with the longest including five nodes originating at SKN7. There are several other 4-node chains that originate at CIN5, MAC1, PHD1, SKN7, and YAP1. Finally, there are two feedforward motifs involving CIN5, ROX1, and YAP6 and SKN7, YAP1, and ROX1. More information about this network can be found in Dahlquist et al. (2015).
- The networks with colored edges shown in the bottom panel display the results of a mathematical model, called GRNmap, where the expression levels of the individual transcription factors were modeled using mass balance ordinary differential equations with a sigmoidal production function. Each equation in the model included a production rate, a degradation rate, weights that denote the magnitude and type of influence of the connected transcription factors (activation or repression), and a threshold of expression. The differential equation model was fit to published yeast cold shock microarray data from Schade et al. (2004) using a penalized nonlinear least squares approach. Model predictions fit the experimental data well, within the 95% confidence interval.
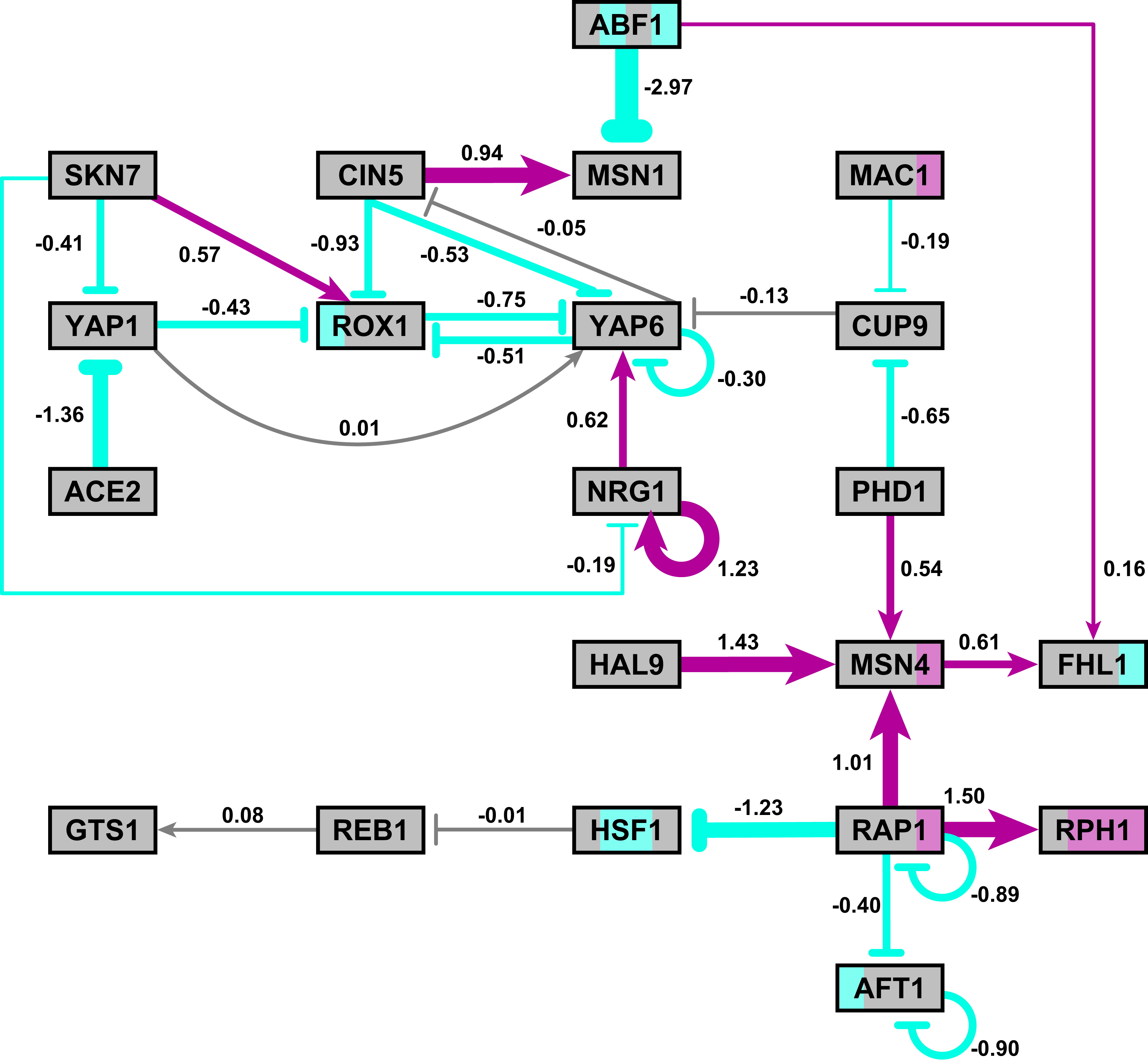
- In particular, GRNsight is displaying the results of the optimized weight parameters. Positive weights > 0 represent an activation relationship and are shown by regular (pointy) arrowheads. One example is that CIN5 activates the expression of MSN1. Negative weights < 0 represent a repression relationship and are shown by a blunt arrowhead (a line segment perpendicular to the edge direction). One example is that ABF1 represses the expression of MSN1.
- The thicknesses of the edges also vary based on the magnitude of the absolute value of the weight. Larger magnitudes have thicker edges and smaller magnitudes have thinner edges. The way that GRNsight determines the edge thickness is as follows. GRNsight divides all weight values by the absolute value of the maximum weight in the matrix to normalize all the values to between zero and 1. GRNsight then adjusts the thickness of the lines to vary continuously from the minimum thickness (for normalized weights near zero) to maximum thickness (normalized weights of 1). For example, in the bottom panel, the thickest edge corresponds to the repression of the expression of MSN1 by ABF1 because it has the highest magnitude weight parameter of -2.97.
- The color of the edge also imparts information about the regulatory relationship. Edges with positive normalized weight values from 0.05 to 1 are colored magenta (red in GRNsight >= v3.0.0). There are 10 magenta edges in this example. Edges with negative normalized weight values from -0.05 to -1 are colored cyan (blue in GRNsight >= v3.0.0). There are 16 cyan edges in this example. Edges with normalized weight values between -0.05 and 0.05 are colored gray to emphasize that their normalized magnitude is near zero and that they have a weak influence on the target gene (5 edges in this example).
- Because of this visualization of the weight parameters, one can make some interesting observations about the behavior of the network. The expression of several genes is controlled by a balance of activation and repression by different regulators. For example, the expression of MSN1 is strongly activated by CIN5, but even more strongly repressed by ABF1. Furthermore, some transcription factors themselves act both as activators of some targets and repressors of other targets. For example, RAP1 activates the expression of MSN4 and RPH1, but represses the expression of AFT1, HSF1, and itself. RAP1 is known to act as both an activator and a repressor. Thus, GRNsight enables one to interpret the weight parameters more easily than one could from the adjacency matrix alone. For more information about the interpretation of the model results, see Dahlquist et al. (2015).
- Note that the nodes in the bottom right panel of the figure are also colored, based on the time course of expression of that gene in the Schade et al. (2004) microarray data (stripes from left to right, 10, 30, and 120 minutes of cold shock, with magenta representing a significant increase in expression relative to the control at time 0, cyan representing a significant decrease in expression relative to the control, and gray representing no significant change in expression relative to the control). This feature is available in GRNsight >= v3.0.0, and described in the section below.
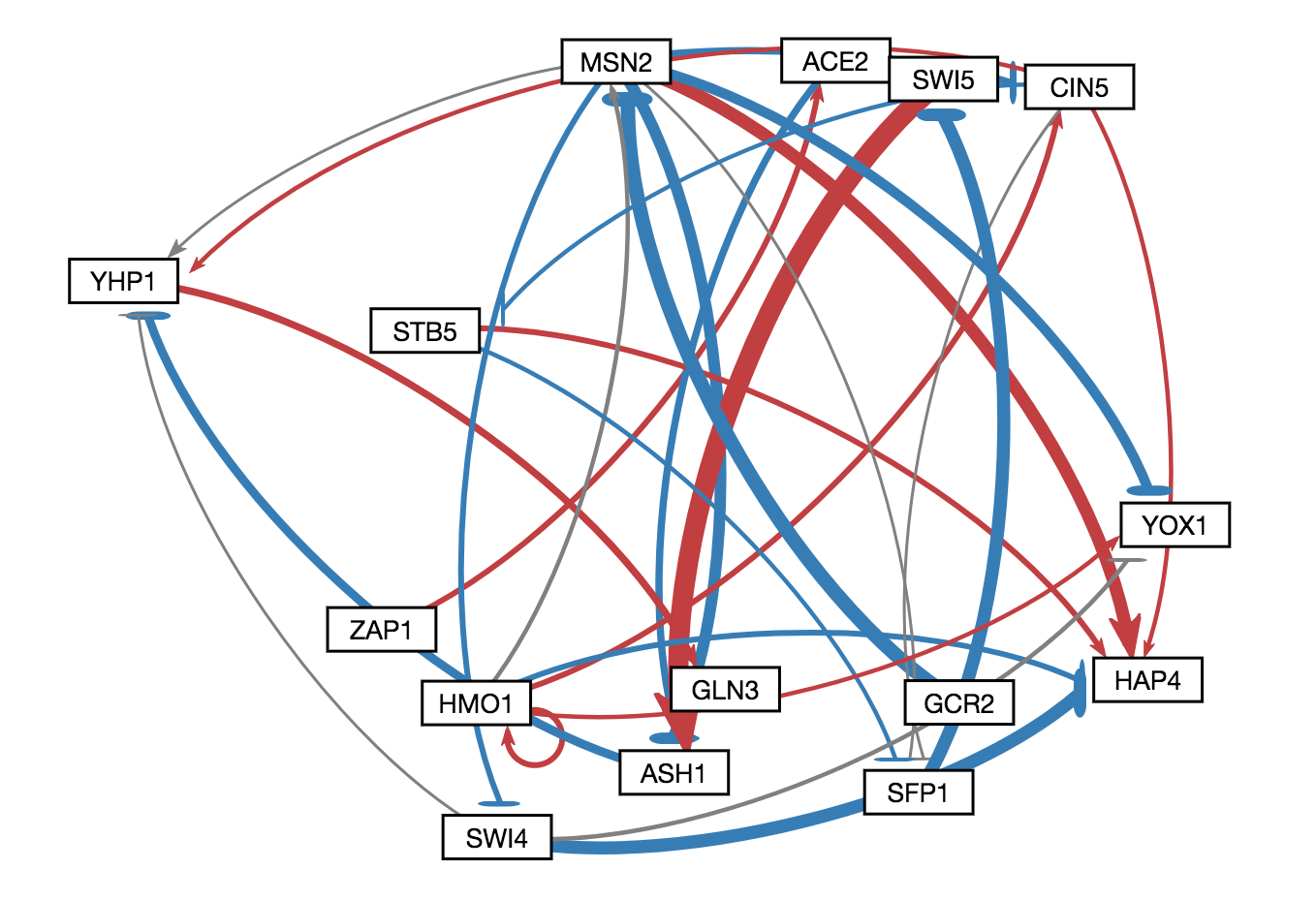
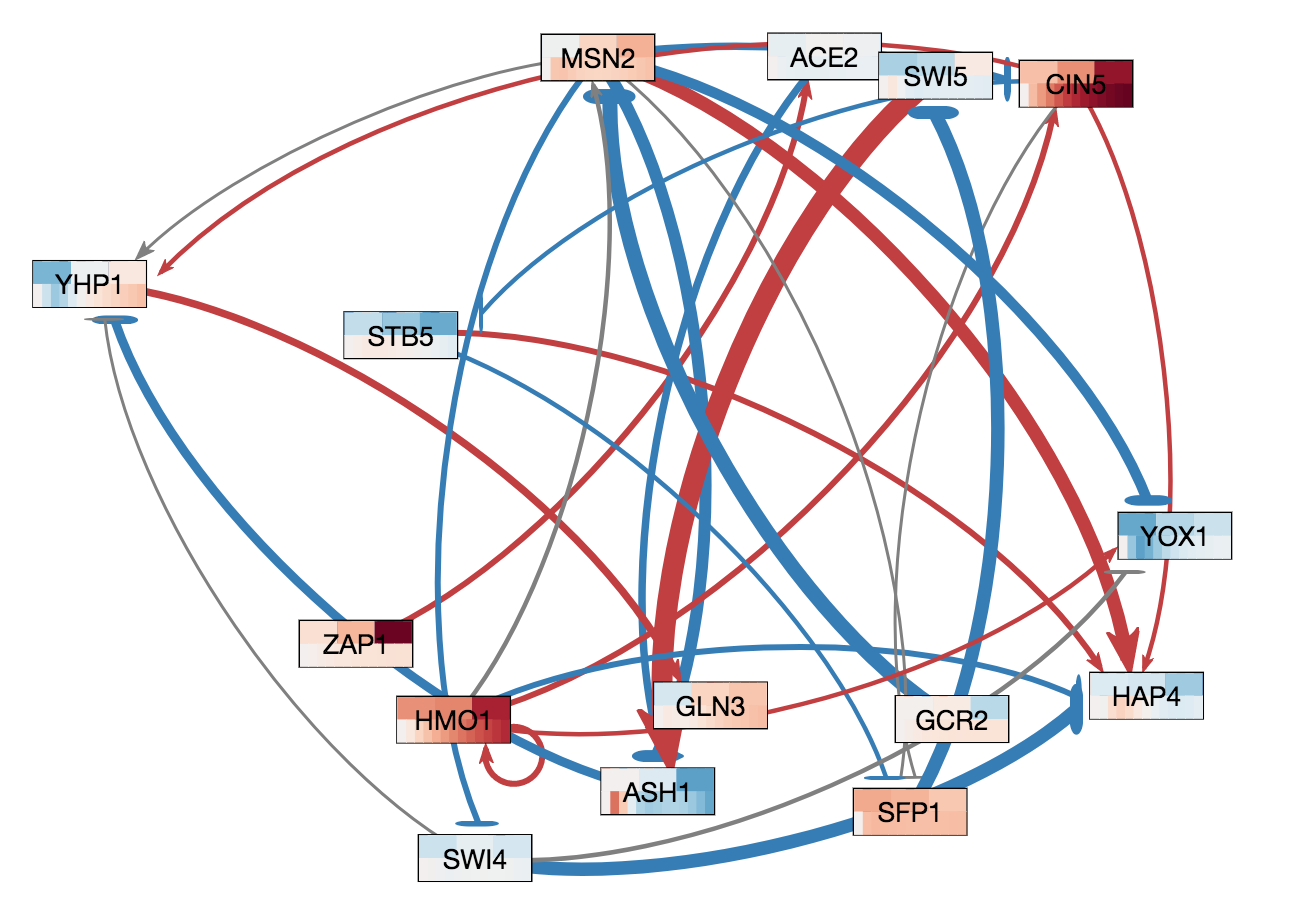
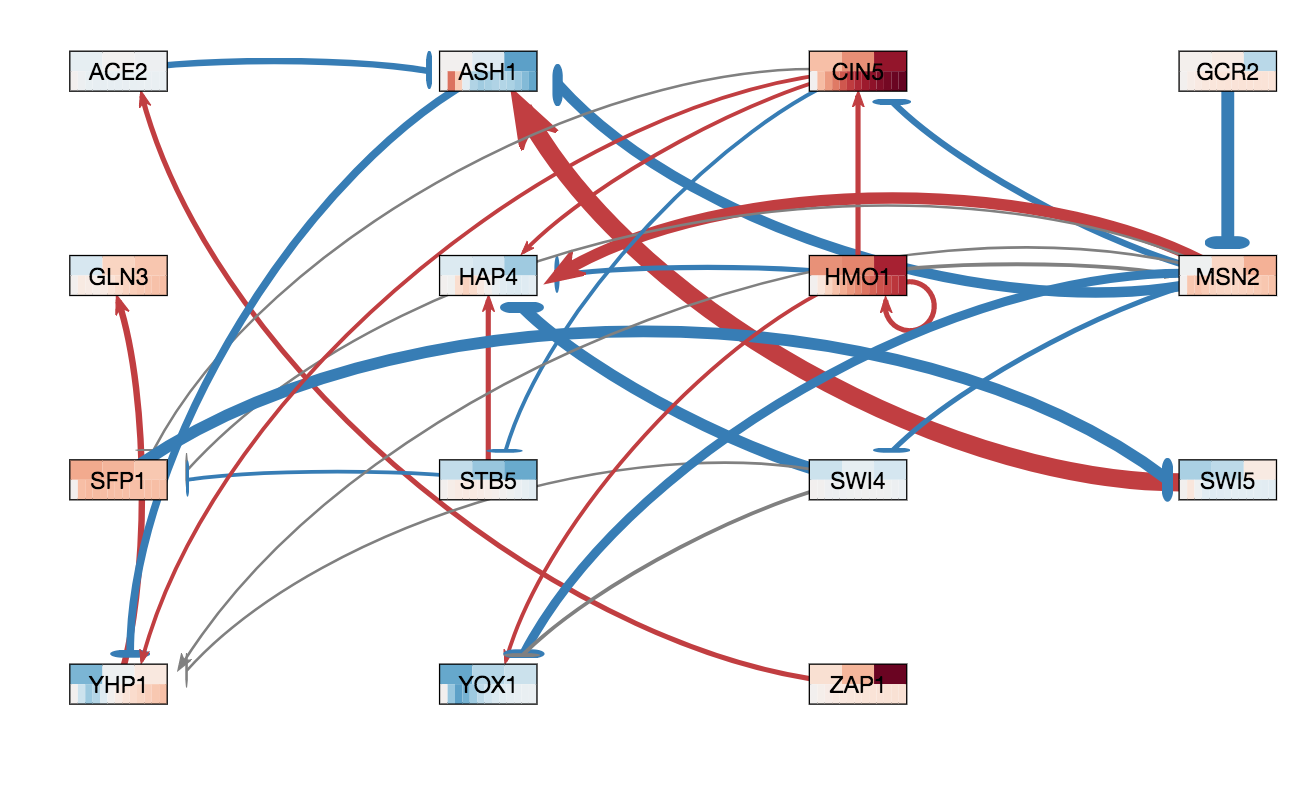
In GRNsight v3.0.0, two major features were added to GRNsight: node coloring visualization for time series expression data and a grid layout option. The following three figures were derived from Demo #2 Weighted GRN (15 genes, 28 edges, Dahlquist Lab unpublished data) using GRNsight v3.0.0. The figure on the left shows an example of the automated force graph layout generated by GRNsight with the new edge coloring scheme introduced in GRNsight v3.0.0. The figure in the middle shows an example of the automated force graph layout with the node coloring feature turned on. The figure on the right shows an example of the grid layout option with the node coloring feature turned on.
 |
 |
 |
- As shown in the figure on the left, the color scheme for edges was changed in GRNsight v3.0.0 to red and blue from magenta and cyan, respectively. The color scheme was changed to provide consistency with the node coloring visualization and to provide more visual contrast over the original coloring scheme for color-blind users.
- Demo #2 Weighted GRN (15 genes, 28 edges, Dahlquist Lab unpublished data) contains expression sheets formatted for node coloring. Nodes in the graph are colored in the middle and left figure to visualize the level of expression of transcription factors over time. Two different datasets are selected for the top and bottom nodes to facilitate visual comparision between the expression values in the datasets. The top half of the nodes are colored using data from the sheet "wt_log2_expression" of Demo #2 and the bottom half of the nodes are colored using data from the sheet "wt_log2_optimized_expression" of Demo #2. The number of vertical node coloring slices in each half of the node correspond to the number of data points in the data set. The color of each vertical slice correspond to the color that the expression value at that time point maps to on the node coloring color scale, located in the sidebar menu of the user interface. By default, -3 maps to the darkest blue shade, while 3 maps to the darkest red shade.
- The figure on the right shows an example of the grid layout option. Grid layout was introduced in GRNsight v3.0.0 to provide an alternative graph layout option to the force graph layout. In grid layout mode, nodes are alphabetically organized in a grid pattern.
In GRNsight v3.1.0, the gene pages feature was added. To access the gene pages, right-click on the text of any given node when a graph is loaded. A page should load similar to the one seen in the leftmost figure below, similar to the one in the browser. Meanwhile, a "page is loading" message should appear, as shown in the figure on the left.
 |
 |
 |
- The figure in the center shows the gene page when it is loaded. The gene name is displayed on the left. he species that the data is collected from is displayed on the GRNSight. Presently, the gene page pulls data from the Uniprot, NCBI Gene, Ensembl, JASPAR, and Saccharomyces Geneome databases.
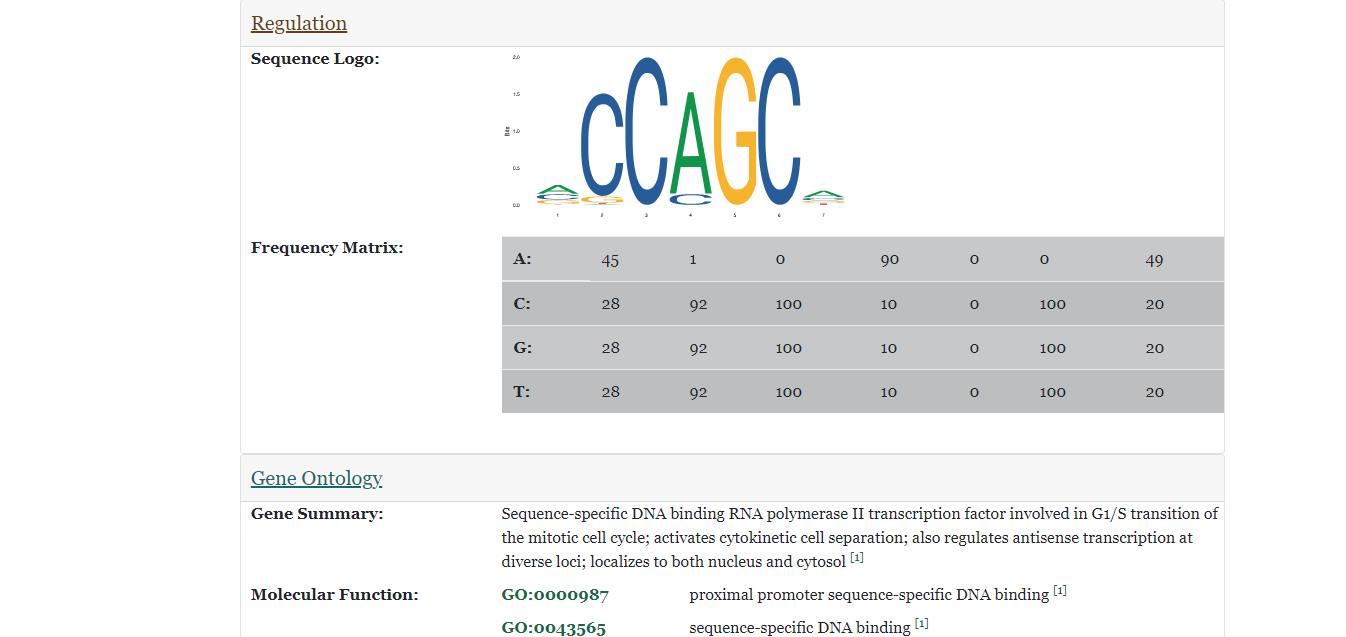
- The figure on the right displays that after clicking on a button corresponding to a certain section, the page is directed to that section and displays information about the specified information about the gene in the species. In this instance, a sequence logo, frequency matrix, and a list of gene ontology values are displayed.
In GRNsight v5.0.0, the species feature was added. GRNsight now supports the following species: “Arabidopsis thaliana”, “Caenorhabditis elegans”, “Drosophila melanogaster”, “Homo sapiens”, “Mus musculus”, and “Saccharomyces cerevisiae.”
- If you don’t add a supported species to the “optimization_parameters” sheet, then it will default to the species “Saccharomyces cerevisiae” and the taxon id “559292.”
- You can change the species in the GRNsight web application under Node > Current Species.
-
To export your data to SIF format, select the menu option Export > Export Data > To Unweighted SIF or File > Export Data > To Weighted SIF. A dialog box will open for you to Save your file. Clicking the Cancel button will return you to the GRNsight web page.
- When an unweighted network is loaded, exporting to an unweighted SIF file will be the only option available.
- When a weighted network is loaded, you have the option to export to an unweighted or weighted SIF file. The difference between the unweighted and weighted SIF file is described below.
-
While there are several valid ways that a SIF file can be formatted for import into GRNsight (Section 1b-i.c), GRNsight follows a particular convention when exporting to SIF.
- GRNsight will export the network data as a tab-delimited text file (not space delimited) with a .sif file extension.
- The suggested filename for export will default to the same filename as was opened/imported with the file extension changed to .sif.
- If you are exporting a weighted network as an unweighted SIF file, the text "_unweighted" will be appended to the suggested filename before the file extension to denote this.
- For unweighted networks, the relationship type “pd” will be used. The relationship type of “pd” is used because it commonly means protein -> DNA (e.g. a regulatory transcription factor binding upstream of a target gene) in the systems biology community. This default value cannot be changed from within GRNsight, but the exported SIF file could be edited in another program to do so.
- For weighted networks, if the user has selected the option of exporting the data as an unweighted network, the relationship type “pd” will be used.
- If the user has selected the option to export a weighted network as a weighted network, the numerical weight value for each edge is given as the relationship type.
- In each case, each line will consist of source node, relationship type, and target node, separated by tab characters. If a particular source node has multiple targets, they will be listed on separate lines. If a particular source node has no targets, it will still be listed on a separate line. The screenshots shown below are examples of how GRNsight exports Demo #3: Unweighted GRN" and Demo #4: Weighted GRN", respectively, to SIF files. (The screenshots display a tab-delimited text file as it would be displayed in Microsoft Excel.) Follow the links to download the sample SIF files.


- Note that these “weighted” SIF files are able to be imported in to Cytoscape (tested with v3.4.0), but the weight values will not be properly stored as edge attributes by Cytoscape because it does not use the SIF format in this way.
- One benefit from this export behavior is that GRNsight can be used to convert network data from an adjacency matrix (in the native GRNsight .xlsx format as described in Section 1b-i.a) to a list of binary interactions (as a SIF file).
- To export your data to GraphML format, select the menu option Export > Export Data > To Unweighted GraphML or File > Export Data > To Weighted GraphML. A dialog box will open for you to Save your file. Clicking the Cancel button will return you to the GRNsight web page.
- When an unweighted network is loaded, exporting to an unweighted GraphML file will be the only option available.
- When a weighted network is loaded, you have the option to export to an unweighted or weighted GraphML file.
- The unweighted and weighted GraphML files will be formatted according to the description in Section 1b-i.d.
- To export your data to Excel format, select the menu option Export > Export Data > To Excel. A selector modal should appear allowing you to set the specifications of your workbook file before exporting it. Once specifications are selected, a dialog box will open for you to Save your workbook. Clicking the Cancel button will return you to the GRNsight web page.
- Selector Modal:
- You can select the workbooks expression data source. You can choose to export expression data from the original workbook or you can choose from data sources within the Expression Database.
- From there you can select which particular network sheets and expression sheets you would like to export from the chosen expression data source or expand upon the workbook by selecting particular additional sheets from the selector modal. By default all available network sheets, expression sheets, and additional sheets are selected to be exported into the workbook file.
- The unweighted and weighted Excel workbooks will be formatted according to the description in Section 1b-i.a.
- To export your data as an image, select the menu option Export > Export Image > To [PNG, SVG, PDF]. A dialog box will appear, allowing you to save your file. Selecting Cancel will return you to the GRNsight web page.
- Notes: If you plan to view an SVG file in Adobe Illustrator, follow these steps:
- Download the graph as an SVG.
- Open the file in a web browser such as Chrome.
- Export it as a PDF from the browser.
- Open the PDF in Adobe Illustrator to view the vector graphic properly.
To print a graph, select the menu item File > Print. Mac users can utilize the native print to PDF function available from their operating system to print the graph to a PDF file. Windows users will need to have the full version of Adobe Acrobat (or other "print to PDF" utility) to print graphs to a PDF file. The Print menu item is disabled until a gene regulatory network graph is loaded. There is a known bug when GRNsight is used with Firefox browsers where the side of the graph gets cut off. This is an issue with Firefox that we cannot solve at this time. There are two workarounds. First, in Firefox, go to the menu item File > Page Setup and check the box Shrink to fit Page Width. Second, you can use the Chrome browser to print to PDF.
For additional documentation, please see the GRNsight Wiki.
To request support or features or report bugs, please submit an issue to GRNsight @ GitHub (requires free GitHub account).